
Spatial dynamics of chromosome translocations in living cells. Solution structure of DAPI selectively bound in the minor groove of a DNA T.T mismatch-containing site: NMR and molecular dynamics studies. Proliferation, cell cycle and apoptosis in cancer. Cellular senescence: when bad things happen to good cells. Temporal and spatial control of cyclin B1 destruction in metaphase. Direct visualization of cell division using high-resolution imaging of M-phase of the cell cycle. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell cycle markers for live cell analyses. Nuclear dynamics of PCNA in DNA replication and repair. Dynamics of DNA replication factories in living cells. Dynamic recruitment of licensing factor Cdt1 to sites of DNA damage. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Estimation of nuclear DNA content in plants using flow cytometry. Automated determination of DNA using the fluorochrome Hoechst 33258. Profiling of DNA replication timing in unsynchronized cell populations. A novel method based on click chemistry, which overcomes limitations of cell cycle analysis by classical determination of BrdU incorporation, allowing multiplex antibody staining. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Measurement of DNA content using propidium iodide (PI) staining of fixed whole cells. Analyzing DNA replication I: labeling animals, tissues, and cells with bromodeoxyuridine (BrdU). Analysis of cellular DNA content by flow cytometry. Hallmarks of cancer: the next generation. A long twentieth century of the cell cycle and beyond. Cell cycle, CDKs and cancer: a changing paradigm. This 1–2-d protocol is applicable to adherent cells, and it is adaptable for use with several DNA dyes. We describe and provide the algorithms for two automated image analysis pipelines and the derivation of cell cycle profiles with both commercial and open-source software.
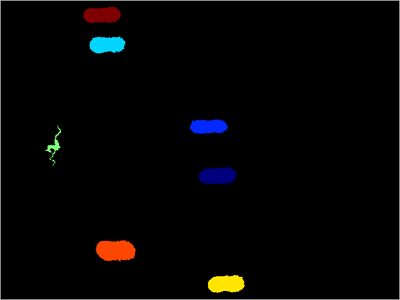
The approach is based on the accurate quantification by image analysis of the integrated nuclear intensity of cells stained with a DNA dye, and it can be used in combination with several histochemical methods. We describe a protocol, suitable for use in high-resolution imaging approaches, for determining cell cycle staging of individual cells by measuring their DNA content by fluorescence microscopy. Studies of the cell cycle have traditionally relied on the analysis of populations, and they often require specific markers or the use of genetically modified systems, making it difficult to determine the cell cycle stage of individual, unperturbed cells. Progression through the cell cycle is one of the most fundamental features of cells.


 0 kommentar(er)
0 kommentar(er)
